User Tools
Sidebar
mm:parameters:manganese:hhewawa
Table of Contents
Manganese(II) - HHEWaWa
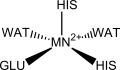
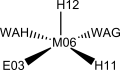
Homoprotocatechuate-2,3-deoxygenase is shown to have five ligands bonded to the iron(II) ion: a glutamate, two histidines and two waters in a square pyramidal geometry. The ligands in manganese(II) deoxygenases are likely to be the same. Other studies show, however, that it might be hexacoordinated with three solvent molecules occupying ligand positions.
Structure chosen to parameterize
| TEST PROTEIN | |
| Protein | Homoprotocatechuate 2,3-Dioxygenase |
| PDB Code | 1F1U |
| Crystallographic Resolution | 1.50 Å |
| Organism | Arthrobacter globiformis |
| [Vetting, 2004] | |
Parameters Determined
Atom Types
Bond Parameters
| BOND | Kl / kcal mol-1 Å-2 | l / Å | REFERENCE |
| M1-NI | 81.53 | 2.1862 | [Neves, 2013] |
| M1-NJ | 78.93 | 2.1763 | |
| M1-E1 | 90.87 | 2.0648 | |
| M1-W1 | 43.45 | 2.2506 | |
| M1-W2 | 45.59 | 2.2237 | |
| NI-CR | 488.0 | 1.335 | |
| NI-CV | 410.0 | 1.394 | |
| NJ-CR | 488.0 | 1.335 | |
| NJ-CV | 410.0 | 1.394 | |
| E1-C | 656.0 | 1.25 | |
| W1-HM | 553.0 | 0.9572 | |
| W2-HM | 553.0 | 0.9572 |
Angle Parameters
| ANGLE | Kθ / kcal mol-1 rad-2 | θ / deg | REFERENCE |
| NI-M1-NJ | 35.670 | 106.2009 | [Neves, 2013] |
| NI-M1-E1 | 66.76 | 90.0029 | |
| NI-M1-W1 | 27.91 | 89.1764 | |
| NI-M1-W2 | 27.25 | 152.3584 | |
| NJ-M1-E1 | 58.53 | 100.4401 | |
| NJ-M1-W1 | 8.378 | 121.3184 | |
| NJ-M1-W2 | 30.649 | 101.1859 | |
| E1-M1-W1 | 8.825 | 136.6245 | |
| E1-M1-W2 | 64.45 | 88.6732 | |
| W1-M1-W2 | 41.88 | 73.2308 | |
| M1-NI-CR | 46.273 | 126.3998 | |
| M1-NI-CV | 46.273 | 126.3998 | |
| M1-NJ-CR | 31.467 | 118.1041 | |
| M1-NJ-CV | 31.467 | 118.1041 | |
| M1-E1-C | 49.84 | 127.7223 | |
| M1-W1-HM | 7.881 | 110.4316 | |
| M1-W2-HM | 26.41 | 109.0419 | |
| NI-CR-NA | 70.0 | 120.0 | |
| NI-CR-H5 | 50.0 | 120.0 | |
| NI-CV-CC | 70.0 | 120.0 | |
| CV-NI-CR | 70.0 | 117.0 | |
| H4-CV-NI | 50.0 | 120.0 | |
| NJ-CR-NA | 70.0 | 120.0 | |
| NJ-CR-H5 | 50.0 | 120.0 | |
| NJ-CV-CC | 70.0 | 120.0 | |
| CV-NJ-CR | 70.0 | 117.0 | |
| H4-CV-NJ | 50.0 | 120.0 | |
| D1-C-CT | 70.0 | 117.0 | |
| D1-C-O2 | 80.0 | 126.0 | |
| E1-C-CT | 70.0 | 117.0 | |
| E1-C-O2 | 80.0 | 126.0 | |
| HM-W1-HM | 100.0 | 104.52 | |
| HM-W2-HM | 100.0 | 104.52 |
Van der Waals Parameters
| ATOM TYPE | Ri / Å | εi / kcal mol-1 | REFERENCE |
| M1 | 1.4544 | 0.03 | [Babu, 2006] |
| NI | 1.824 | 0.17 | |
| NJ | 1.824 | 0.17 | |
| E1 | 1.6612 | 0.21 | |
| W1 | 1.7683 | 0.152 | |
| W2 | 1.7683 | 0.152 | |
| HM | 0.6000 | 0.0157 | [Cornell, 1995] |
Validation of Parameters from MD Simulations
Bond Parameters
| BOND | l0 crystal | l0 opt | ‹l›MD ± σ(l) | ‹l›MD ± 2σ(l) |
| M1-NI | 2.21 | 2.19 | 2.15 ± 0.06 | [2.086; 2.205] |
| M1-NJ | 2.03 | 2.18 | 2.15 ± 0.06 | [2.086; 2.212] |
| M1-E1 | 2.11 | 2.06 | 2.08 ± 0.06 | [2.017; 2.133] |
| M1-W1 | 2.38 | 2.25 | 2.18 ± 0.08 | [2.098; 2.266] |
| M1-W2 | 2.18 | 2.22 | 2.17 ± 0.08 | [2.095; 2.255] |
Angle Parameters
| ANGLE | θ0 crystal | θ0 QM | ‹θ›MD ± σ(θ) | ‹θ›MD ± 2σ(θ) |
| NI-M1-NJ | 95.6 | 106.2 | 110 ± 4 | [102.1; 117.0] |
| NI-M1-E1 | 86.7 | 90.0 | 94 ± 3 | [86.9; 100.5] |
| NI-M1-W1 | 91.4 | 89.2 | 80 ± 4 | [71.5; 89.3] |
| NI-M1-W2 | 151.4 | 152.4 | 152 ± 4 | [143.9; 159.7] |
| NJ-M1-E1 | 97.3 | 100.4 | 102 ± 4 | [93.6; 110.1] |
| NJ-M1-W1 | 93.4 | 121.3 | 122 ± 7 | [108.5; 135.2] |
| NJ-M1-W2 | 109.2 | 101.2 | 97 ± 4 | [88.6; 104.7] |
| E1-M1-W1 | 169.3 | 136.6 | 135 ± 7 | [121.3; 148.7] |
| E1-M1-W2 | 103.5 | 88.7 | 89 ± 4 | [82.0; 96.6] |
| W1-M1-W2 | 73.6 | 73.2 | 78 ± 4 | [69.5; 86.8] |
Downloads
| Parameter File | .frcmod |
| Coordination Sphere Charges | .off |
| Homoprotocatechuate-2,3-dioxygenase’s PDB File (1F1U.pdb) LEAPRC file | .pdb, leaprc |
References
Neves, R.P.P.; Sousa, S.F.; Fernandes, P.A.; Ramos, M.J.. Parameters for Molecular Dynamics Simulations of Manganese-Containing Metalloproteins . J. Chem. Theory Comput., 2013, 9 (6), 2718–2732
mm/parameters/manganese/hhewawa.txt · Last modified: 2013/10/03 18:58 by ruineves
Except where otherwise noted, content on this wiki is licensed under the following license: CC Attribution-Share Alike 3.0 Unported