User Tools
Sidebar
mm:parameters:manganese:denwawawa
Table of Contents
Manganese(II) - DENWaWaWa
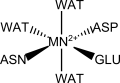
D-Glutarate Dehydratase is analogous to mandelate racemase and enolase. The manganese coordination sphere has a manganese(II) atom bonded to a glutamate, an aspartate and an asparagine from the same subunit.
Structure chosen to parameterize
| TEST PROTEIN | |
| Protein | D-Glucarate Dehydratase |
| PDB Code | 3NXL |
| Crystallographic Resolution | 1.89 Å |
| Organism | Burkholderia sp. 383 |
| [Fedorov, 2010] | |
Parameters Determined
Atom Types
Bond Parameters
| BOND | Kl / kcal mol-1 Å-2 | l / Å | REFERENCE |
| M1-NW | 35.45 | 2.2737 | [Neves, 2013] |
| M1-D1 | 67.93 | 2.1478 | |
| M1-E1 | 92.09 | 2.0644 | |
| M1-W1 | 50.25 | 2.2190 | |
| M1-W2 | 33.63 | 2.3313 | |
| M1-W3 | 41.73 | 2.2662 | |
| NW-C | 570.0 | 1.229 | |
| D1-C | 656.0 | 1.25 | |
| E1-C | 656.0 | 1.25 | |
| W1-HM | 553.0 | 0.9572 | |
| W2-HM | 553.0 | 0.9572 | |
| W3-HM | 553.0 | 0.9572 |
Angle Parameters
| ANGLE | Kθ / kcal mol-1 rad-2 | θ / deg | REFERENCE |
| NW-M1-D1 | 50.31 | 168.8916 | [Neves, 2013] |
| NW-M1-E1 | 36.14 | 92.7969 | |
| NW-M1-W1 | 38.04 | 100.6537 | |
| NW-M1-W2 | 30.57 | 95.9566 | |
| NW-M1-W3 | 33.91 | 78.7883 | |
| D1-M1-E1 | 35.463 | 100.5525 | |
| D1-M1-W1 | 64.49 | 85.2832 | |
| D1-M1-W2 | 48.3 | 73.9644 | |
| D1-M1-W3 | 32.46 | 94.4909 | |
| E1-M1-W1 | 27.16 | 94.9567 | |
| E1-M1-W2 | 16.85 | 166.1705 | |
| E1-M1-W3 | 44.90 | 89.8914 | |
| W1-M1-W2 | 13.16 | 97.1854 | |
| W1-M1-W3 | 26.160 | 175.1204 | |
| W2-M1-W3 | 23.22 | 78.0892 | |
| M1-NW-C | 10.785 | 144.0557 | |
| M1-D1-C | 47.557 | 125.7501 | |
| M1-E1-C | 36.476 | 122.3574 | |
| M1-W1-HM | 24.67 | 106.0283 | |
| M1-W2-HM | 9.470 | 89.9559 | |
| M1-W3-HM | 23.78 | 97.5290 | |
| HM-W1-HM | 100.0 | 104.52 | |
| HM-W2-HM | 100.0 | 104.52 | |
| HM-W3-HM | 100.0 | 104.52 | |
| NW-C-CT | 80.0 | 120.40 | |
| NW-C-N | 80.0 | 122.90 | |
| D1-C-CT | 70.0 | 117.0 | |
| D1-C-O2 | 80.0 | 126.0 | |
| E1-C-CT | 70.0 | 117.0 | |
| E1-C-O2 | 80.0 | 126.0 |
Van der Waals Parameters
| ATOM TYPE | Ri / Å | εi / kcal mol-1 | REFERENCE |
| M1 | 1.4544 | 0.03 | [Babu, 2006] |
| NW | 1.6612 | 0.21 | |
| D1 | 1.6612 | 0.21 | |
| E1 | 1.6612 | 0.21 | |
| W1 | 1.7683 | 0.152 | |
| W2 | 1.7683 | 0.152 | |
| W3 | 1.7683 | 0.152 | |
| HM | 0.6000 | 0.0157 | [Cornell, 1995] |
Validation of Parameters from MD Simulations
Bond Parameters
| BOND | l0 crystal | l0 opt | ‹l›MD ± σ(l) | ‹l›MD ± 2σ(l) |
| M1-D1 | 2.28 | 2.15 | 2.16 ± 0.06 | [2.095; 2.224] |
| M1-E1 | 2.45 | 2.06 | 2.13 ± 0.06 | [2.066; 2.186] |
| M1-NW | 2.40 | 2.27 | 2.29 ± 0.09 | [2.195; 2.383] |
| M1-W1 | 2.26 | 2.22 | 2.21 ± 0.08 | [2.127; 2.290] |
| M1-W2 | 2.51 | 2.33 | 2.3 ± 0.1 | [2.222; 2.416] |
| M1-W3 | 2.47 | 2.27 | 2.25 ± 0.09 | [2.165; 2.341] |
Angle Parameters
| ANGLE | θ0 crystal | θ0 QM | ‹θ›MD ± σ(θ) | ‹θ›MD ± 2σ(θ) |
| D1-M1-E1 | 100.8 | 100.6 | 98 ± 4 | [89.3; 106.1] |
| D1-M1-NW | 154.1 | 168.9 | 165 ± 3 | [158.6; 172.1] |
| D1-M1-W1 | 84.5 | 85.3 | 85 ± 4 | [78.1; 92.1] |
| D1-M1-W2 | 93.4 | 73.9 | 70 ± 4 | [61.8; 77.6] |
| D1-M1-W3 | 80.9 | 94.5 | 91 ± 4 | [82.4; 100.0] |
| E1-M1-NW | 66.7 | 88.4 | 94 ± 4 | [85.9; 102.0] |
| E1-M1-W1 | 113.1 | 94.9 | 96 ± 5 | [86.4; 104.6] |
| E1-M1-W2 | 160.1 | 166.2 | 157 ± 5 | [148.0; 166.1] |
| E1-M1-W3 | 95.6 | 89.9 | 88 ± 5 | [78.7; 97.4] |
| NW-M1-W1 | 95.7 | 100.7 | 101 ± 4 | [92.7; 109.3] |
| NW-M1-W2 | 86.6 | 95.9 | 97 ± 4 | [88.2; 105.4] |
| NW-M1-W3 | 98.7 | 78.8 | 82 ± 4 | [73.2; 90.6] |
| W1-M1-W2 | 107.8 | 97.2 | 101 ± 5 | [91.3; 111.6] |
| W1-M1-W3 | 149.8 | 175.1 | 172 ± 4 | [164.7; 179.6] |
| W2-M1-W3 | 69.2 | 78.1 | 74 ± 6 | [62.6; 85.8] |
Downloads
| Parameter File | .frcmod |
| Coordination Sphere Charges | .off |
| D-Glutarate Dehydratase’s PDB File (3NXL.pdb) LEAPRC file | .pdb, leaprc |
References
Neves, R.P.P.; Sousa, S.F.; Fernandes, P.A.; Ramos, M.J.. Parameters for Molecular Dynamics Simulations of Manganese-Containing Metalloproteins . J. Chem. Theory Comput., 2013, 9 (6), 2718–2732
mm/parameters/manganese/denwawawa.txt · Last modified: 2013/10/03 18:53 by ruineves
Except where otherwise noted, content on this wiki is licensed under the following license: CC Attribution-Share Alike 3.0 Unported