User Tools
Sidebar
mm:parameters:manganese:ed_py2_wawa
This is an old revision of the document!
Table of Contents
Manganese(II) - ED[PY2]WaWa
| Coordination Sphere | Oxidation State: Mn(II) Spin Multiplicity: 6 |
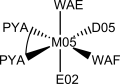
 |  1) 1) |
Structure chosen to parameterize
| TEST PROTEIN | |
| Protein | Pyruvate Kinase |
| PDB Code | 2G50 |
| Crystallographic Resolution | 1.65 Å |
| Organism | Oryctolagus cuniculus |
| [Williams, 2006] | |
Parameters Determined
Atom Types
Bond Parameters
| BOND | Kl / kcal mol-1 Å-2 | l / Å | REFERENCE |
| M1-D1 | 71.90 | 2.0926 | [Neves, 2013] |
| M1-E1 | 68.77 | 2.1399 | |
| M1-R1 | 55.39 | 2.1382 | |
| M1-R2 | 16.78 | 2.4332 | |
| M1-W1 | 24.75 | 2.3873 | |
| M1-W2 | 50.08 | 2.2004 | |
| D1-C | 656.0 | 1.25 | |
| E1-C | 656.0 | 1.25 | |
| W1-HM | 553.0 | 0.9572 | |
| W2-HM | 553.0 | 0.9572 | |
| c-R2 | 648.0 | 1.214 | |
| c-R1 | 648.0 | 1.214 |
Angle Parameters
| ANGLE | K? / kcal mol-1 rad-2 | ? / deg | REFERENCE |
| D1-M1-E1 | 38.27 | 96.6209 | [Neves, 2013] |
| D1-M1-R1 | 23.41 | 104.8054 | |
| D1-M1-R2 | 12.445 | 169.3708 | |
| D1-M1-W1 | 69.3 | 72.8299 | |
| D1-M1-W2 | 26.19 | 103.5245 | |
| E1-M1-R1 | 29.696 | 97.7449 | |
| E1-M1-R2 | 13.94 | 93.9317 | |
| E1-M1-W1 | 25.05 | 168.0717 | |
| E1-M1-W2 | 52.2 | 89.0644 | |
| R1-M1-R2 | 128.3 | 80.3387 | |
| R1-M1-W1 | 20.47 | 80.0896 | |
| R1-M1-W2 | 18.05 | 149.8517 | |
| R2-M1-W1 | 13.736 | 96.5418 | |
| R2-M1-W2 | 27.09 | 77.9339 | |
| W1-M1-W2 | 15.008 | 98.6963 | |
| M1-D1-C | 14.829 | 125.0463 | |
| M1-E1-C | 42.056 | 122.8295 | |
| M1-R1-c | 47.47 | 122.0775 | |
| M1-R2-c | 57.66 | 110.2798 | |
| M1-W1-HM | 12.10 | 86.3993 | |
| M1-W2-HM | 28.96 | 88.1336 | |
| HM-W1-HM | 100.0 | 104.52 | |
| HM-W2-HM | 100.0 | 104.52 | |
| HM-W3-HM | 100.0 | 104.52 | |
| D1-C-CT | 70.0 | 117.0 | |
| D1-C-O2 | 80.0 | 126.0 | |
| E1-C-CT | 70.0 | 117.0 | |
| E1-C-O2 | 80.0 | 126.0 | |
| c3-c-R2 | 68.0 | 123.11 | |
| R1-c-c | 66.8 | 122.34 | |
| o-c-R1 | 79.1 | 127.33 | |
| c-c-R2 | 66.8 | 122.34 |
Van der Waals Parameters
| ATOM TYPE | Ri / Å | ?i / kcal mol-1 | REFERENCE |
| M1 | 1.4544 | 0.03 | [Babu, 2006] |
| D1 | 1.6612 | 0.21 | |
| E1 | 1.6612 | 0.21 | |
| R1 | 1.6612 | 0.2100 | |
| R2 | 1.6612 | 0.2100 | |
| W1 | 1.7683 | 0.152 | |
| W2 | 1.7683 | 0.152 | |
| HM | 0.6000 | 0.0157 | [Cornel, 1995] |
Validation of Parameters from MD Simulations
Bond Parameters
| BOND | l0 crystal | l0 opt | ‹l›MD ± ?(l) | ‹l›MD ± 2?(l) |
| M1-D1 | 1.97 | 2.09 | 2.16 ± 0.06 | [2.095; 2.225] |
| M1-E1 | 1.96 | 2.14 | 2.22 ± 0.06 | [2.153; 2.281] |
| M1-R1 | 1.98 | 2.14 | 2.13 ± 0.07 | [2.059; 2.192] |
| M1-R2 | 1.95 | 2.43 | 2.4 ± 0.1 | [2.276; 2.473] |
| M1-W1 | 2.29 | 2.39 | 2.4 ± 0.1 | [2.289; 2.537] |
| M1-W2 | 2.26 | 2.20 | 2.18 ± 0.08 | [2.100; 2.258] |
Angle Parameters
| ANGLE | ?0 crystal | ?0 QM | ‹?›MD ± ?(?) | ‹?›MD ± 2?(?) |
| D1-M1-E1 | 97.0 | 96.6 | 99 ± 5 | [89.0; 108.1] |
| D1-M1-R1 | 90.8 | 104.8 | 103 ± 5 | [93.1; 112.4] |
| D1-M1-R2 | 153.6 | 169.4 | 170 ± 5 | [159.4; 180.1] |
| D1-M1-W1 | 78.1 | 72.8 | 75 ± 4 | [67.8; 82.2] |
| D1-M1-W2 | 113.2 | 103.5 | 110 ± 5 | [99.5; 120.6] |
| E1-M1-R1 | 91.7 | 97.7 | 101 ± 5 | [91.1; 110.3] |
| E1-M1-R2 | 105.0 | 93.9 | 75 ± 5 | [64.3; 86.1] |
| E1-M1-W1 | 163.9 | 168.1 | 168 ± 4 | [159.5; 176.5] |
| E1-M1-W2 | 84.8 | 89.1 | 80 ± 4 | [71.4; 87.7] |
| R1-M1-R2 | 74.4 | 80.3 | 75 ± 2 | [70.9; 79.1] |
| R1-M1-W1 | 103.6 | 80.1 | 79 ± 7 | [63.7; 93.7] |
| R1-M1-W2 | 156.0 | 149.9 | 146 ± 5 | [136.7; 155.4] |
| R2-M1-W1 | 84.2 | 96.5 | 111 ± 7 | [97.0; 124.2] |
| R2-M1-W2 | 83.5 | 77.9 | 73 ± 5 | [63.3; 82.5] |
| W1-M1-W2 | 83.1 | 98.7 | 105 ± 10 | [85.6; 123.9] |
Downloads
| Parameter File | .frcmod |
| Coordination Sphere Charges | .off |
| ENZYME’s PDB File (PDB_CODE.pdb)\ LEAPRC file | .pdb, leaprc |
References
Neves, R.P.P.; Sousa, S.F.; Fernandes, P.A.; Ramos, M.J.. Parameters for Molecular Dynamics Simulations of Manganese-Containing Metalloproteins . J. Chem. Theory Comput., 2013, 9 (6), 2718–2732
1)
The schemes represent geometrical dispositions for crystal lattices. These should be roughly similar to the corresponding distorted geometries in enzymatic coordination spheres.
mm/parameters/manganese/ed_py2_wawa.1374822824.txt.gz · Last modified: 2013/07/26 08:13 by ruineves
Except where otherwise noted, content on this wiki is licensed under the following license: CC Attribution-Share Alike 3.0 Unported