User Tools
Sidebar
mm:parameters:manganese:hhdh_ho_o
Table of Contents
Manganese(III) - HHDH[HO]O
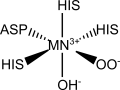
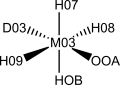
Electronic spectroscopy studies on the resting and enzyme-substrates states of Manganese Superoxide Dismutase support the hypothesis of a side-on peroxo complex with the superoxide anion. The superoxide anion coordinates to the manganese ion between two histidines. The spin multiplicity of the coordination sphere has been reported as a quartet, due to an antiferromagnetic coupling between the superoxide and manganese species.
Structure chosen to parameterize
| TEST PROTEIN | |
| Protein | Manganese Superoxide Dismutase |
| PDB Code | 1MNG |
| Crystallographic Resolution | 1.80 Å |
| Organism | Thermus thermophilus |
| [Lah, 1995] | |
Parameters Determined
Atom Types
Bond Parameters
| BOND | Kl / kcal mol-1 Å-2 | l / Å | REFERENCE |
| M1-NI | 82.11 | 2.1879 | [Neves, 2013] |
| M1-NJ | 123.46 | 2.0161 | |
| M1-NK | 138.95 | 2.0175 | |
| M1-D1 | 111.54 | 2.0630 | |
| M1-R1 | 96.51 | 1.9361 | |
| M1-W1 | 227.9 | 1.7993 | |
| NI-CR | 488.0 | 1.335 | |
| NI-CV | 410.0 | 1.394 | |
| NJ-CR | 488.0 | 1.335 | |
| NJ-CV | 410.0 | 1.394 | |
| NK-CR | 488.0 | 1.335 | |
| NK-CV | 410.0 | 1.394 | |
| D1-C | 656.0 | 1.25 | |
| R1-O | 384.3 | 1.430 | |
| W1-HM | 553.0 | 0.9572 |
Angle Parameters
| ANGLE | Kθ / kcal mol-1 rad-2 | θ / deg | REFERENCE |
| NI-M1-NJ | 93.41 | 91.0663 | [Neves, 2013] |
| NI-M1-NK | 96.11 | 86.5034 | |
| NI-M1-D1 | 118.5 | 82.0566 | |
| NI-M1-W1 | 77.66 | 176.5931 | |
| NI-M1-R1 | 83.7 | 84.2181 | |
| NJ-M1-NK | 97.22 | 172.0561 | |
| NJ-M1-D1 | 74.40 | 93.5533 | |
| NJ-M1-W1 | 96.31 | 90.3808 | |
| NJ-M1-R1 | 95.80 | 86.9561 | |
| NK-M1-D1 | 75.579 | 93.5990 | |
| NK-M1-W1 | 92.14 | 92.4642 | |
| NK-M1-R1 | 99.09 | 85.2759 | |
| D1-M1-W1 | 83.35 | 94.7758 | |
| D1-M1-R1 | 70.04 | 166.2720 | |
| W1-M1-R1 | 61.59 | 98.9401 | |
| M1-NI-CR | 53.671 | 126.4370 | |
| M1-NI-CV | 53.671 | 126.4370 | |
| M1-NJ-CR | 40.495 | 123.3813 | |
| M1-NJ-CV | 40.495 | 123.3813 | |
| M1-NK-CR | 31.220 | 112.3780 | |
| M1-NK-CV | 31.220 | 112.3780 | |
| M1-D1-C | 47.094 | 135.5495 | |
| M1-W1-HM | 36.67 | 106.2284 | |
| M1-R1-O | 41.32 | 118.4913 | |
| NI-CR-NA | 70.0 | 120.0 | |
| NI-CR-H5 | 50.0 | 120.0 | |
| NI-CV-CC | 70.0 | 120.0 | |
| CV-NI-CR | 70.0 | 117.0 | |
| H4-CV-NI | 50.0 | 120.0 | |
| NJ-CR-NA | 70.0 | 120.0 | |
| NJ-CR-H5 | 50.0 | 120.0 | |
| NJ-CV-CC | 70.0 | 120.0 | |
| CV-NJ-CR | 70.0 | 117.0 | |
| H4-CV-NJ | 50.0 | 120.0 | |
| NK-CR-NA | 70.0 | 120.0 | |
| NK-CR-H5 | 50.0 | 120.0 | |
| NK-CV-CC | 70.0 | 120.0 | |
| CV-NK-CR | 70.0 | 117.0 | |
| H4-CV-NK | 50.0 | 120.0 | |
| D1-C-CT | 70.0 | 117.0 | |
| D1-C-O2 | 80.0 | 126.0 |
Van der Waals Parameters
| ATOM TYPE | Ri / Å | εi / kcal mol-1 | REFERENCE |
| M1 | 1.4544 | 0.03 | [Babu, 2006] |
| NI | 1.824 | 0.17 | |
| NJ | 1.824 | 0.17 | |
| NK | 1.824 | 0.17 | |
| D1 | 1.6612 | 0.21 | |
| R1 | 1.6612 | 0.2100 | |
| O | 1.6612 | 0.2100 | |
| W1 | 1.7210 | 0.2104 | |
| HM | 0.6000 | 0.0157 | [Cornell, 1995] |
Validation of Parameters from MD Simulations
Bond Parameters
| BOND | l0 crystal | l0 opt | ‹l›MD ± σ(l) | ‹l›MD ± 2σ(l) |
| M1-D1 | 1.80 | 2.06 | 2.09 ± 0.05 | [2.038; 2.142] |
| M1-NI | 2.15 | 2.19 | 2.20 ± 0.06 | [2.141; 2.254] |
| M1-NJ | 2.12 | 2.02 | 2.02 ± 0.05 | [1.969; 2.063] |
| M1-NK | 2.16 | 2.02 | 2.05 ± 0.05 | [2.003; 2.094] |
| M1-W1 | 2.07 | 1.80 | 1.79 ± 0.04 | [1.752; 1.825] |
| M1-R1 | 2.68 | 1.94 | 1.93 ± 0.06 | [1.872; 1.984] |
Angle Parameters
| ANGLE | θ0 crystal | θ0 QM | ‹θ›MD ± σ(θ) | ‹θ›MD ± 2σ(θ) |
| NI-M1-NJ | 92.4 | 91.1 | 92 ± 2 | [86.9; 96.6] |
| NI-M1-NK | 90.3 | 86.5 | 89 ± 2 | [84.3; 93.7] |
| NI-M1-D1 | 89.1 | 82.1 | 82 ± 2 | [77.6; 87.0] |
| NI-M1-W1 | 175.5 | 176.6 | 176 ± 2 | [172.3; 179.7] |
| NI-M1-R1 | 97.6 | 84.2 | 86 ± 2 | [80.8; 90.8] |
| NJ-M1-NK | 130.8 | 172.1 | 170 ± 2 | [165.7; 174.9] |
| NJ-M1-D1 | 110.1 | 93.6 | 93 ± 3 | [87.0; 98.6] |
| NJ-M1-W1 | 90.0 | 90.4 | 87 ± 2 | [82.6; 92.4] |
| NJ-M1-R1 | 66.4 | 87.0 | 85 ± 3 | [80.4; 90.5] |
| NK-M1-D1 | 119.2 | 93.6 | 97 ± 3 | [90.9; 102.1] |
| NK-M1-W1 | 91.0 | 92.5 | 92 ± 3 | [86.9; 97.1] |
| NK-M1-R1 | 64.6 | 85.3 | 85 ± 2 | [80.5; 90.2] |
| D1-M1-W1 | 86.5 | 94.8 | 96 ± 3 | [90.5; 101.8] |
| D1-M1-R1 | 172.4 | 166.3 | 167 ± 3 | [162.3; 172.5] |
| W1-M1-R1 | 86.8 | 98.9 | 96 ± 3 | [90.1; 101.3] |
Downloads
| Parameter File | .frcmod |
| Coordination Sphere Charges | .off |
| Manganese Superoxide Dismutase’s PDB File (1MNG.pdb) LEAPRC file | .pdb, leaprc |
References
Neves, R.P.P.; Sousa, S.F.; Fernandes, P.A.; Ramos, M.J.. Parameters for Molecular Dynamics Simulations of Manganese-Containing Metalloproteins . J. Chem. Theory Comput., 2013, 9 (6), 2718–2732
mm/parameters/manganese/hhdh_ho_o.txt · Last modified: 2013/10/03 18:56 by ruineves
Except where otherwise noted, content on this wiki is licensed under the following license: CC Attribution-Share Alike 3.0 Unported