User Tools
Sidebar
mm:parameters:iron:cccc
This is an old revision of the document!
Table of Contents
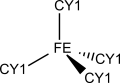
Iron(III) - CCCC
| Coordination Sphere | Oxidation State: Fe(III) Spin Multiplicity: 6 |
 | PATH TO IMAGE2 1) |
Structure chosen to parameterize
| TEST PROTEIN | |
| Protein | Rubredoxin |
| PDB Code | 1IRO |
| Crystallographic Resolution | 1.10 Å |
| Organism | Clostridium pasteurianum |
| [Dauter, 1996] | |
Parameters Determined
Atom Types
Bond Parameters
| BOND | Kl / kcal mol-1 Å-2 | l / Å | REFERENCE |
| FE-SF | 80.75 | 2.3170 | [Carvalho, 2013] |
| CT-SF | 237.00 | 1.8100 |
Angle Parameters
| ANGLE | Kθ / kcal mol-1 rad-2 | θ / deg | REFERENCE |
| SF-FE-SF | 33.15 | 109.8650 | [Carvalho, 2013] |
| CT-SF-FE | 27.60 | 100.9150 | |
| SF-CT-H1 | 50.00 | 109.5000 | |
| CT-CT-SF | 50.00 | 108.6000 |
Van der Waals Parameters
| ATOM TYPE | Ri / Å | εi / kcal mol-1 | REFERENCE |
| FE | 2.6500 | 0.0450 | [Giammona, 1984] |
| SF | 2.0000 | 0.2500 |
Downloads
| Parameter File | .frcmod |
| Coordination Sphere Charges | .off |
| Rubredoxin’s PDB File (1IRO.pdb)\ LEAPRC file | .pdb, leaprc |
References
A.T.P. Carvalho, A.F.S. Teixeira, MJ.Ramos. Parameters for Molecular Dynamics Simulations of Iron-Sulfur Proteins. J. Comput. Chem., 2013, 34, 1540-1548
D. A. Giammona, Ph.D. Thesis; University of California: Davis, 1984
1)
The schemes represent geometrical dispositions for crystal lattices. These should be roughly similar to the corresponding distorted geometries in enzymatic coordination spheres.
mm/parameters/iron/cccc.1375724876.txt.gz · Last modified: 2013/08/05 18:47 by ruineves
Except where otherwise noted, content on this wiki is licensed under the following license: CC Attribution-Share Alike 3.0 Unported