User Tools
Sidebar
This is an old revision of the document!
Table of Contents
Copper(II)/Zinc(II) - HHH[Cu]H[Zn]HHD
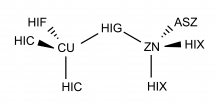
The metal center of Superoxide Dismutase is hereby presented, in which the zinc atom is bonded to 3 histidines, and 1 aspartate (monodentate) and the copper atom to 4 histidines. Validation for this metal centre in the protein environment is additionally provided.
| Coordination Sphere | Oxidation state: Cu(II) / Zn(II) Spin multiplicity: 1 |
 |  1) 1) |
Structure chosen to parameterize
| TEST PROTEIN | |
| Protein | Superoxide Dismutase |
| PDB Code | 1CBJ |
| Crystallographic Resolution | 1.65 Å |
| Organism | Bos taurus |
| [Hough, 1999] | |
Parameters Determined
Atom Types
Bond Parameters
| BOND | Kl / kcal mol-1 Å-2 | l / Å | REFERENCE |
| ZN-NB | 96.7 | 2.046 | [Branco, 2006] |
| ZN-OS | 92.0 | 1.949 | |
| CU-NB | 101.1 | 2.022 |
Angles Parameters
| ANGLE | Kθ / kcal mol-1 rad-2 | θ / deg | REFERENCE |
| NB-CU-NB | 24.6 | 147.19 | [Branco, 2006] |
| NB-ZN-NB | 44.0 | 113.53 | |
| NB-ZN-OS | 21.7 | 111.75 |
Van der Waals Parameters
| ATOM TYPE | Ri / Å | εi / kcal mol-1 | REFERENCE |
| ZN | [Ryde, 1995] | ||
| CU | [Ryde, 1995] | ||
| OS | 1.6612 | 0.2100 | |
| NB | 1.8240 | 0.1700 |
Validation of Parameters from MD Simulations
Bond Parameters
The validation considering two SOD structures was conducted: 1 for the fully reduced enzyme - 1Q0E (resolution of 1.15 Å); and for the structure used in the parameterization - 1CBJ. The two chains were considered in the validation (A, and B), and the brackets denotes the decoordinated residue upon reduction.
| BOND / Å | l0 crystal 1CBJ:A | l0 crystal 1CBJ:B | l0 crystal 1Q0E:A | l0 crystal 1Q0E:B | ‹l›MD ± 0.06 A | ‹l›MD ± 0.06 B |
| CU-NB(H44) | 2.07 | 2.00 | 2.02 | 2.05 | 2.03 | 2.04 |
| CU-NB(H46) | 2.00 | 2.17 | 1.96 | 2.01 | 2.08 | 2.07 |
| CU-NB(H61) | (3.19) | 2.20 | (3.39) | (3.32) | 2.03 | 2.03 |
| CU-NB(H118) | 2.03 | 2.19 | 2.08 | 2.04 | 2.03 | 2.03 |
| ZN-NB(H61) | 1.97 | 2.02 | 2.05 | 2.05 | 2.03 | 2.03 |
| ZN-NB(H69) | 2.03 | 2.07 | 2.05 | 2.05 | 2.05 | 2.04 |
| ZN-NB(H78) | 2.07 | 1.75 | 2.06 | 2.06 | 2.06 | 2.05 |
| ZN-OS(D81) | 1.62 | 1.84 | 1.99 | 2.00 | 1.90 | 1.92 |
Angle Parameters
| ANGLE | θ0 crystal 1CBJ:A | θ0 crystal 1CBJ:B | ‹θ›MD ± 5 A | ‹θ›MD ± 5 B |
| H44-CU-H46 | 140.9 | 138.2 | 133.0 | 133.8 |
| H44-CU-H61 | 72.5 | 86.8 | 96.6 | 97.3 |
| H44-CU-H118 | 101.1 | 93.0 | 98.7 | 98.0 |
| H46-CU-H61 | 92.5 | 97.6 | 101.2 | 101.5 |
| H46-CU-H118 | 117.6 | 99.7 | 107.5 | 106.6 |
| H61-CU-H118 | 118.5 | 153.9 | 121.6 | 121.4 |
| H61-ZN-H69 | 108.1 | 110.0 | 106.5 | 105.1 |
| H61-ZN-H78 | 110.0 | 110.2 | 106.7 | 106.3 |
| H61-ZN-A81 | 104.6 | 103.9 | 111.4 | 112.4 |
| H69-ZN-H78 | 118.9 | 117.3 | 112.8 | 112.9 |
| H69-ZN-D81 | 98.0 | 99.7 | 105.3 | 103.8 |
| H78-ZN-D81 | 115.9 | 114.5 | 113.4 | 115.5 |
Downloads
References
Branco, J.F.B.; Fernandes, P.A.; Ramos, M.J.. Molecular Dynamics Simulations of the Enzyme Cu, Zn Superoxide Dismutase . J. Phys. Chem. B, 2006, 110, 16754-16762