User Tools
Sidebar
This is an old revision of the document!
Table of Contents
MADAMM
Operating systems: MacOS and Linux.
Contacts : Nuno Sousa Cerqueira Pedro A. Fernandes Maria João Ramos
Citation :
VsLab—An implementation for virtual high-throughput screening using AutoDock and VMD.
N. M. F. S. A. Cerqueira, J. Ribeiro, P. A. Fernandes AND M. J. Ramos
International Journal of Quantum Chemistry, 2011, 111(6):1208–1212
DOI:10.1002/qua.22738
1. Introduction
Computational capacity has increased dramatically over the last decade making possible the use of more sophisticated and computationally intensive methods in computer-assisted drug design. However, dealing with receptor flexibility in docking methodologies is still a thorny issue. The main reason behind this difficulty is the large number of degrees of freedom that have to be considered in this kind of calculations. However, neglecting it, leads to poor docking results in terms of binding pose prediction in real-world settings.
In order to overcome these limitations we present an automated procedure called MADAMM that allows flexibilization of both the receptor and the ligand during a Multi stAged Docking with an Automated Molecular Modeling protocol. Generally speaking the software uses standard docking software and molecular mechanics force fields in the core process and a set of scripts that automates the process without the intervention of the user. In order to simplify the use of MADAMM a graphical interface has also been developed.
The results obtained with this methodology show that this protocol can lead to dramatic improvements in both sampling and scoring over conventional single rigid protein docking. We observe that the orientation of particular residues, at the interface between the protein and the ligand, have a crucial influence on the way they interact.
At the moment the results indicate that MADAMM can be viewed as a powerful tool for investigating ligand binding poses, allowing the researcher to understand the importance of protein flexibility during the binding processes of the ligands. Moreover this program can be viewed as a valuable tool to predict the binding of ligands in receptors where no experimental data is available.
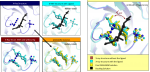
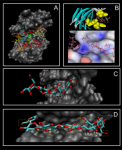
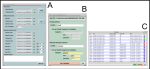
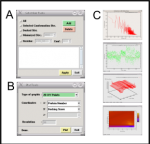
2. ScreenShots
3. Download
In order to run the vsÇab plug-in, the Visual Molecular dynamics software (VMD) must be installed. It can be found at http://www.ks.uiuc.edu/Research/vmd/ .
- VsLab Plug-in : v1.0
4. Installation
VsLab can only be installed in LINUX and MAC operating systems, due to the limitations of the GOLD and Charmm software.
To install MADAMM just download the zip file and extract it using the following command:
tar -xvzf MADAMM.tar.gz
After the extraction go to the directory MADAMM and run the file:
cd MADAMM ./MADAMM.sh
In order to run MADAMM software you also need to install dependencies. Please go to Install Dependencies for more information.
4.1. System Dependencies
Two programs must be installed in order to run vsLab:
- The program VMD (http://www.ks.uiuc.edu/Research/vmd )
- The sqlite database engine (http://www.sqlite.org)
- AutoDock Suite (http://autodock.scripps.edu)
The first program can be easily installed with the tutorials available in the VMD site ( version 1.8.7 is required).
The sqlite database engine is generally installed in all UNIX/LINUX systems (to see if it is installed just write in the terminal: sqlite) If it is not installed you can install writing the following commands in the terminal under the root privileges:
Ubuntu OS : sudo apt-get install sqlite fedora OS : yum install sqlite Mac : (see user manual)
VsLab also depends on autodock and autogrid executables. Therefore they must be installed in the systems before the installation of vLab proceeds.
The installation of AutoDockTools program is also required.
4.2. Install VsLab
Only three steps are required to run VsLab:
- Unordered List ItemExtract the file vsLab.tar.gz to a directory of your choice.
tar -xvzf vsLab.tar.gz
- Run the program install.tcl and follow the instructions:
cd vsLab tclsh install.tcl
Then the user just have to follow the guidelines that are provided by the script.
After the installation ends you can find vsLab inside VMD application, at the plug-in the menu: Extensions» PortoBioComp»virtal screening Lab
5. Tutorial
not available yet.