User Tools
Sidebar
This is an old revision of the document!
Table of Contents
Introduction
Several Density Functional Theory (DFT) benchmarks have been performed by our group. DFT is less demanding than other computational methods with similar accuracy; and is able to include electron correlation in the calculations at a fraction of the time of post-Hartree-Fock methodologies. It also permits the study of molecular systems containing up to 200 atoms, a feature that is not yet feasible with high accuracy methods such as CCSD(T) or even secondorder Möller-Plesset perturbation theory (MP2). Presently, no universally and transferable selection of DFT functionals can be employed, rendering the choice of DFT methodologies, highly influenced by the problem at hand. Consequently, an extensive evaluation of the performance of several density functionals in different fronts (structure, kinetics, thermochemistry, and nonbonded interactions) has emerged in the literature throughout the years, and is crucial when developing a project based on these methodologies.
In our group a review was attained, presenting a brief outline of the density functional theory and of the historic development of the field focusing later on several types of density functionals, and finishing with a detailed analysis of the performance of DFT across a wide range of chemical properties and system types, up to 2006. In addition, benchmark efforts in transition metal chemistry have been described, together with reaction pathways, and proton transfer of aminoacid side-chains in solution.
Review
In this review a brief outline of the density functional theory and of the historic development of the field is provided, focusing later on several types of density functionals (available up to 2006), and it finishes with a detailed analysis of the performance of DFT across a wide range of chemical properties and system types, reviewed from recent benchmarking studies (up to 2006), which encompass several well-established density functionals together with the most recent efforts in the field. Globally, an overall picture of the level of performance of the plethora of currently available density functionals for each chemical property is drawn, with particular attention being dedicated to the relative performance of the popular B3LYP density functional.
Sousa S.F., Fernandes P.A., Ramos M.J. General Performance of Density Functionals - a Review . J. Phys. Chem. A, 2007, 111 (42), 10439-10452
Benchmarks
Transition metals
A set of 44 Zinc-ligand bond-lengths and of 60 ligand-metal-ligand bond angles from 10 diverse transition-metal complexes, representative of the coordination spheres of typical biological Zn systems, were used to evaluate the performance of a total of 18 commonly available density functionals in geometry determination. Five different basis sets were considered for each density functional, namely two all-electron basis sets and three basis sets including popular types of effective-core potentials: Los Alamos, Steven-Basch-Krauss, and Stuttgart-Dresden. For the determination of Zn-ligand bond-lengths and angles, BB1K, MPWB1K, MPW1K, B97-2 and TPSS are suggested as the strongest alternatives within the tested functionals. In addition, the results show that the use of effective-core potentials (in particular Stuttgart-Dresden) has a very limited impact, in terms of accuracy, in the determination of metal-ligand bond-lengths and angles in Zinc-complexes, and is a good and safe alternative to the use of an all electron basis set such as 6-31G(d) or 6-311G(d,p).
Sousa S.F., Carvalho E.S., Ferreira D.M., Tavares I.S., Fernandes P.A., Ramos M.J., and Gomes J.A.N.F. Comparative analysis of the performance of commonly available density functionals in the determination of geometrical parameters for zinc complexes . J. Comput. Chem., 2009, 30 (16), 2752–2763
In the present study, we have compared the performance of the density functional theory (DFT) functionals B1B95, B3LYP, B97-2, BP86, and BPW91 with MP2 for geometry determination in biological mononuclear Zn complexes. A total of 15 different basis sets, of rather diverse complexity, were tested, several which included also three different types of common effective-core potentials. In addition, the ability to describe mononuclear Zn biological systems using relatively simple models of the metal coordination sphere, comprising only the metal atom and a simplified representation of the ligands at the first coordination sphere, starting from a set of high-resolution X-ray crystallographic structures, is evaluated for 90 combinations of method/basis set. The results show that the use of such models allows for a relatively accurate description of the Zn-ligand bond lengths. Globally, B3LYP had the best average performance in the test, closely followed by MP2, whereas B1B95 was the least accurate method. The study also points out B3LYP/CEP-121G and B3LYP/SDD, as the best compromise between accuracy and CPU time for the geometrical characterization of metal-ligand bond lengths in Zn biological systems.
Sousa S.F., Fernandes P.A., Ramos M.J. Comparative Assessment of Theoretical Methods for the Determination of Geometrical Properties in Biological Zinc Complexes . J. Phys. Chem. B, 2007, 111 (30), 9146–9152
In this study, a set of 50 transition-metal complexes of Cu(I) and Cu(II), were used in the evaluation of 18 density functionals in geometry determination. In addition, 14 different basis sets were considered, including four commonly used Pople’s all-electron basis sets; four basis sets including popular types of effective-core potentials: Los Alamos, Steven-Basch-Krauss, and Stuttgart-Dresden; and six triple-zeta basis sets. The results illustrate the performance of different methodological alternatives for the treatment of geometrical properties in relevant copper complexes, pointing out Double-Hybrid (DH) and Long-range Correction (LC) Generalized Gradient Approximation (GGA) methods as better descriptors of the geometry of the evaluated systems. These however, are associated with a computational cost several times higher than some of the other methods employed, such as the M06 functional, which has also demonstrated a comparable performance. In addition, the results show that the use of effective-core potentials has a limited impact, in terms of the accuracy in the determination of metal-ligand bondlengths and angles in our dataset of copper complexes. Hence, these could become a good alternative for the geometrical description of these systems, if one is considering larger copper complexes where the computational cost could be an issue.
Sousa S.F., Pinto G.R.P., Ribeiro A.J.M., Coimbra J.T.S., Fernandes P.A., and Ramos M.J. Comparative Analysis of the Performance of Commonly Available Density Functionals in the Determination of Geometrical Parameters for Copper Complexes . J. Comput. Chem., 2013, 34 (24), 2079–2090
Reaction Pathways
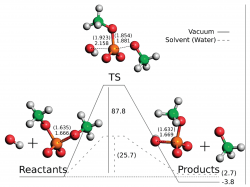
Phosphodiester bonds are an important chemical component of biological systems, and their hydrolysis and formation reactions are involved in major steps throughout metabolic pathways of all organisms. In this work, we applied dimethylphosphate as a model for this kind of bonds and calculated the potential energy surface for its hydrolysis at the approximated CCSD(T)/CBS/B3LYP/6-311++G(2d,2p) level. By varying the nucleophile (water or hydroxide) and the medium (vacuum or aqueous implicit solvent) we obtained and described four reaction paths. These structures were then used in a DFT functional benchmarking in which we tested a total of 52 functionals. Furthermore, the performances of HF, MP2, MP3, MP4, and CCSD were also evaluated. This benchmarking showed that MPWB1K, MPW1B95, and PBE1PBE are the more accurate functionals to calculate the energies of dimethylphosphate hydrolysis as far as activation and reaction energies are concerned. If considering only the activation energies, MPWB1K, MPW1B95, and B1B95 give the lowest errors when comparing to CCSD(T). A basis set benchmarking on the same system shows that 6-311+G(2d,2p) is the best basis set concerning the relationship between computational time and accuracy. We believe that our results will be of great help to further studies on related phosphodiester systems. This includes not only pure chemical problems but also biochemical studies in which DNA, RNA, and phospholipids are required to be depicted at a quantum level.
Ribeiro A.J.M., Ramos M.J., and Fernandes P.A. Benchmarking of DFT Functionals for the Hydrolysis of Phosphodiester Bonds . J. Chem. Theory Comput., 2010, 6 (8), 2281–2292
Proton Transfer
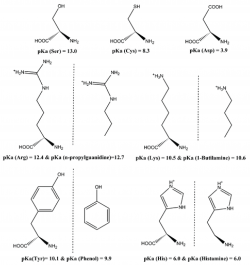
The ionization states of amino acids influence the structure, function, stability, solubility, and reactivity of proteins and are difficult to determine unambiguously by experimental means. Thus, it is very important to be able to determine them theoretically and with high reliability. We have analyzed how well DFT functionals, often used to characterize complex and large models such as proteins, describe the zero-point-exclusive proton affinity at 0 K, PAel0K, for the ionizable side chains of lysine (Lys), histidine (His), arginine (Arg), and aspartate (Asp–) as well as the cysteine (Cys–), serine (Ser–), and tyrosine (Tyr–) anions. The reference values PAel0K were determined at the very accurate CCSD(T)/CBS level. Those values were obtained by the sum of the complete basis set limit of the MP2 energies plus a CCSD(T) correction term evaluated with the aug-cc-pVTZ basis set. The complete basis set limit of MP2 energies was determined using the Truhlar and Helgaker extrapolation schemes. A new, important, and consistent DFT benchmarking database for PAel0K and for proton transfer between two different ionizable side chains, ΔPAel0K, is provided, making this work relevant to all studies with ionizable amino acids side chains that use DFT. Among the 64 density functionals tested, the MPW1B95-D3, XYG3, MPW1B95, B1B95-D3, BMK, BMK-D3, M06-2X, B1LYP, B1B95, PBE1PBE, CAM-B3LYP, B97-1, PBE1KCIS, B3P86, CAM-B3LYP-D3, B3LYP, B98, M06-L, and M06 provide the most accurate PAel0K values for all ionizable amino acids studied, with errors below 1.5 kcal/mol, which translate into an error of less than 1 pKa unit in solution. Furthermore, among the best rated to predict PAel0K, we have found that M06-2X was the most accurate density functional for proton transfers between different amino acids.
Bras, N.F., Perez, M.A.S., Fernandes P.A., Silva, P.J., and Ramos M.J Accuracy of Density Functionals in the Prediction of Electronic Proton Affinities of Amino Acid Side Chains . J. Chem. Theory Comput., 2011, 7 (12), 3898–3908
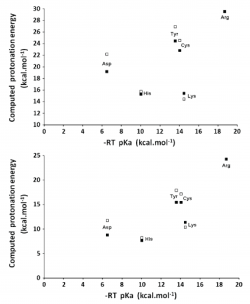
We have studied the influence of implicit solvent models, inclusion of explicit water molecules, inclusion of vibrational effects, and density functionals on the quality of the predicted pKa of small amino acid side chain models. We found that the inclusion of vibrational effects and explicit water molecules is crucial to improve the correlation between the computed and the experimental values. In these micro-solvated systems, the best agreement between DFT-computed electronic energies and benchmark values is afforded by BHHLYP and B97-2. However, approaching experimental results requires the addition of more than three explicit water molecules, which generates new problems related to the presence of multiple minima in the potential energy surface. It thus appears that a satisfactory ab initio prediction of amino acid side chain pKa will require methods that sample the configurational space in the presence of large solvation shells, while at the same time computing vibrational contributions to the enthalpy and entropy of the system under study in all points of that surface. Pending development of efficient algorithms for those computations, we strongly suggest that whenever counterintuitive protonation states are found in a computational study (e.g., the presence of a neutral aspartate/ neutral histidine dyad instead of a deprotonated aspartate/ protonated histidine pair), the reaction profile should be computed under each of the different protonation microstates by constraining the relevant N–H or O–H bonds, in order to avoid artifacts inherent to the complex nature of the factors contributing to the pKa.
Silva, P.J., Perez, M.A.S., Bras, N.F., Fernandes P.A., and Ramos M.J Improving the study of proton transfers between amino acid side chains in solution: choosing appropriate DFT functionals and avoiding hidden pitfalls . Theor. Chem. Acc., 2012, 131 (3), 1179