Software
As part of my research, I have/like to develop scientific software to analyse, process and simulate, both data and models. Here's a list of some of them.
All my programs are public domain and open-source. Contact me if you need assistance installing and/or using one of them. Virtually all of them are associated with a publication. If you have an interesting and challenging problem you can also contact me.
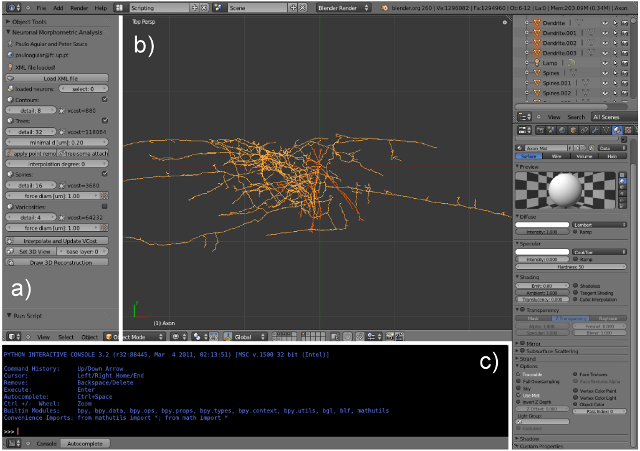
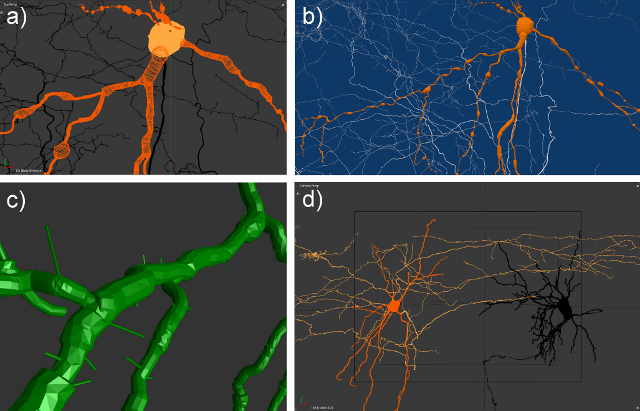
Py3DN
Py3DN, is an open-source solution providing innovative tools to analyze 3D data collected with the widely used commercial system Neurolucida (MBF). It allows the construction of mathematical representations of neuronal topology, visualization and a variety of morphometric analysis on the neuronal structures. Above all, it provides a flexible environment where new types of analyses can be easily set up allowing total freedom to test new hypotheses. The application was developed in Python and uses Blender (an open source program) to produce detailed 3D representations of raw and processed data.
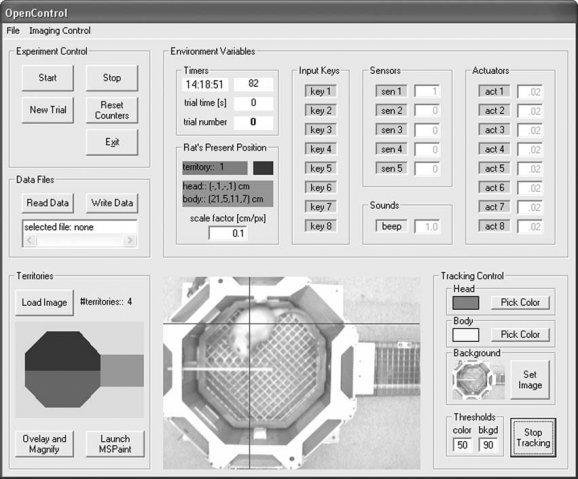
OpenControl
OpenControl is an open-source VBasic software for running fully automated behavioral experiments. It integrates video-tracking of the animal, control of maze actuators from hardware sensors or from online video tracking and recording of experimental data
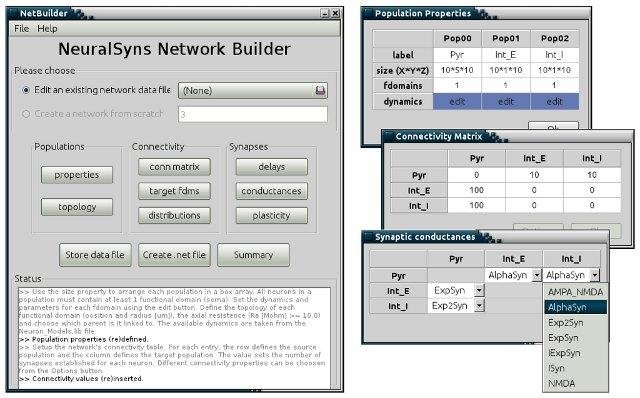
NeuralSyns
NeuralSyns is a neural simulator for large spiking networks. New neuron and synapse dynamics can be added, and complex network models can be easily built using the included tool NetBuilder. OpenGL is used to show the network's architecture and activity
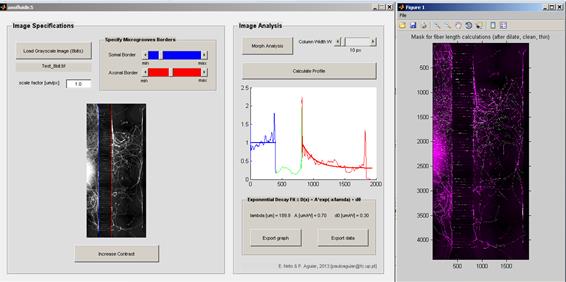
AxoFluidic
The AxoFluidic software was developed with the objective of quantify the axonal outgrowth within compartmentalized microfluidic platforms, where three distinct domains can be easily identified: somal side, microgrooves and axonal side
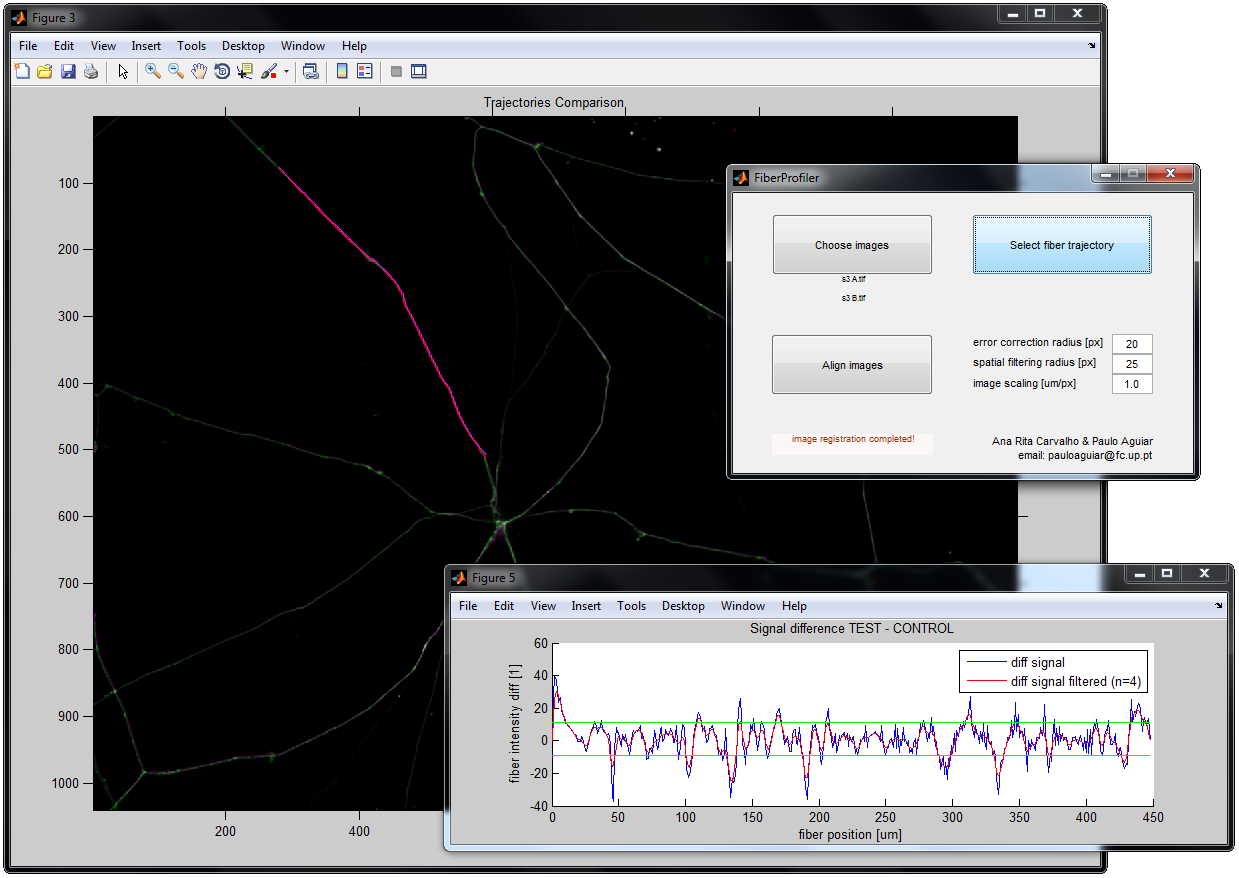
FiberProfiler
FiberProfiler is able to calculate fluorescent intensity profiles along neuronal dendrites/axons, and calculate spatially locked differences in intensity profiles between two time instants.
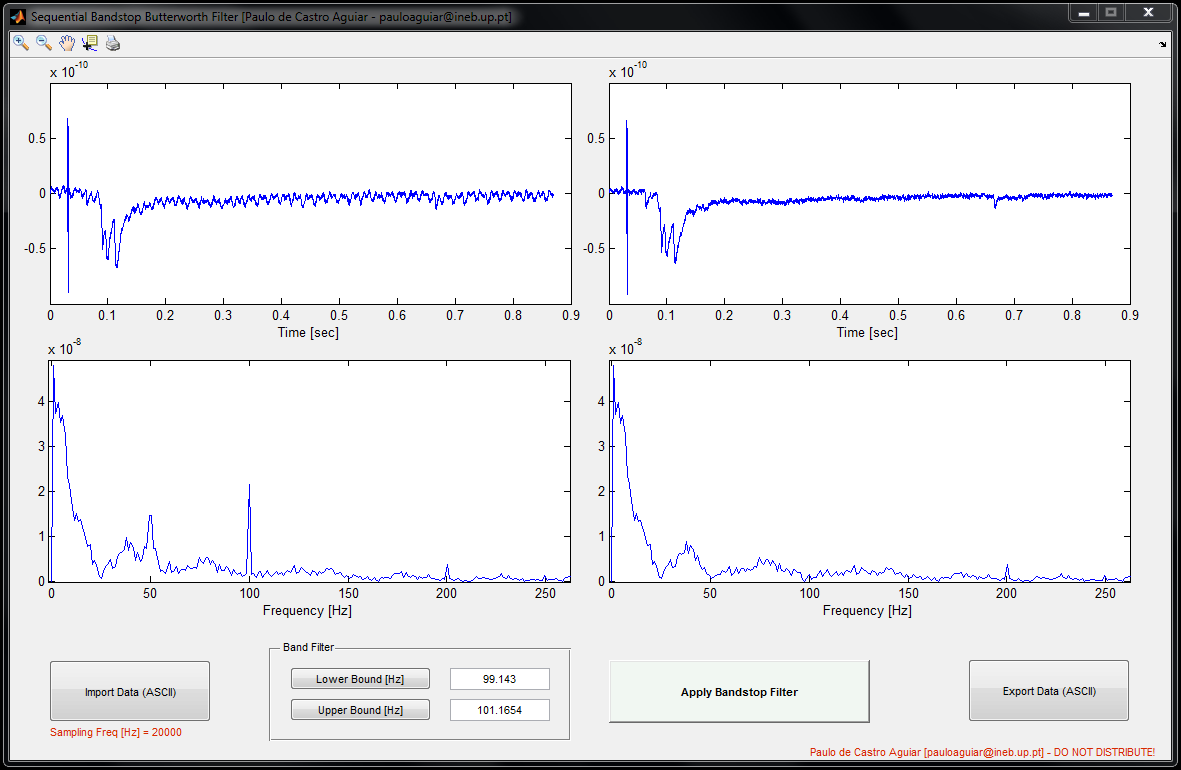
SequentialButterworthBandpassFilter
SequentialButterworthBandpassFilter is a simple tool to remove unwanted frequencies from data samples. We use it in electrophysiology data if we need to filter the 50Hz component. It works however with other types of data.
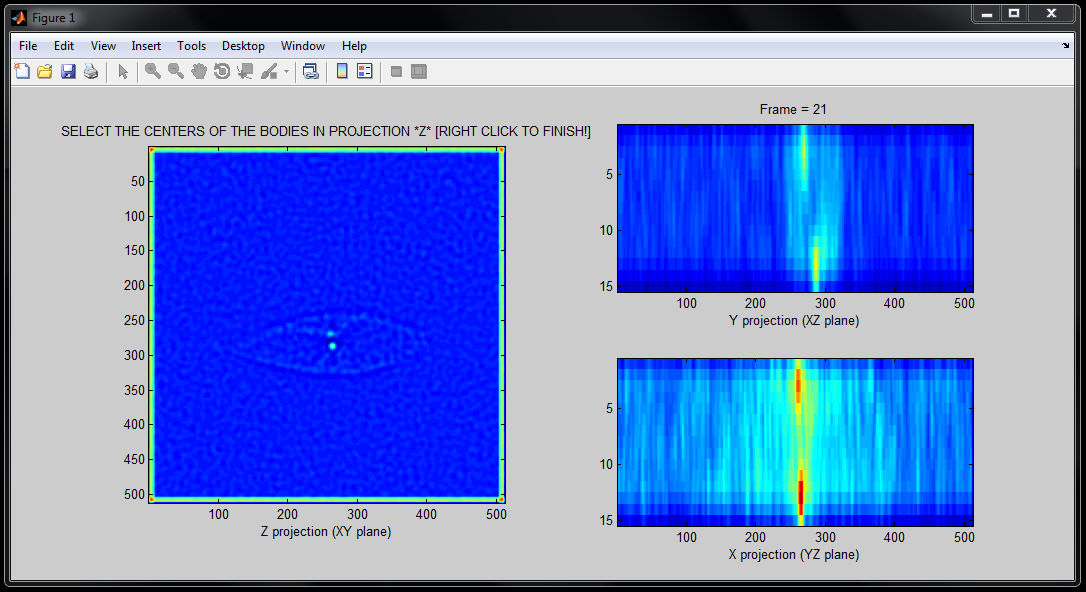
TrackCentrosomes
TrackCentrosomes is a semi-automated tool to track in 4D (x,y,z,t) centrosomes. It is also able to fit an ellipsoid to cell/nuclear membranes.
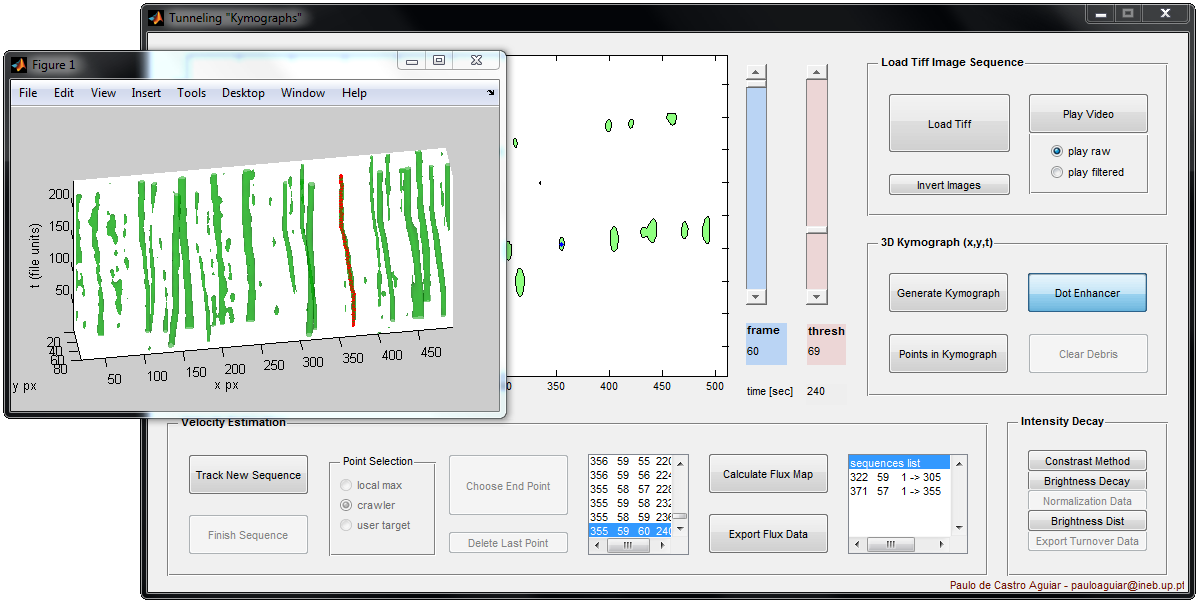
TunnelingKymograph
TunnelingKymograph is used to infer the 2D speed of multiples particles. It is able to calculate flux maps even for sub-pixel velocities (i.e. when the velocity is < 1px/time_step). We use it to calculate the velocity of nanoparticles under axonal transport, and to calculate tubulin flux in the mitotic spindle.